Mathematical models can integrate different data and thus make new, often quantitative, predictions about the behavior of a system beyond pure data analysis. In biological systems, there is an immense biological variability between different datasets, which is difficult to handle in modeling. For example, how a population of genetically identical cells reacts to stimulation is very heterogeneous. This heterogeneity is also present on larger scales, such as in tissues, organs, and organisms.
A Markov process is a stochastic process that is characterized by the fact that it is “memoryless”. The future behavior therefore only depends on the current state of the process and is independent of the past.
The integration of datasets that have a large variability poses a challenge to modeling and often requires stochastic modeling approaches or statistical methods of model calibration that are adapted to the specific problem. Nicole Radde, Professor of Mathematical Modeling and Simulation of Cellular Systems at the University of Stuttgart, and her team are developing such modeling approaches and methods in their basic research. “For example, there’s a standard approach for modeling the stochastic behavior of biochemical reaction networks for smaller numbers of molecules: the Chemical Master Equation, which, however, only works for very small systems,” Nicole Radde explains.
Moments are characteristic quantities of probability distributions. The best-known moments are the expected value, and the variance, which is a measure of how far this random variable deviates from the expected value on average.
The Chemical Master Equation is a system of linear differential equations and is based on the theory of Markov processes. It yields a time-dependent probability distribution over the set of all possible configurations of the system. However, this approach does not work for larger systems, as the set of configurations can usually be very large or even infinitely large, which makes further approximation schemes necessary and leads to very CPU-intensive computations. Obtaining a direct solution of the Chemical Master Equation is often not possible.
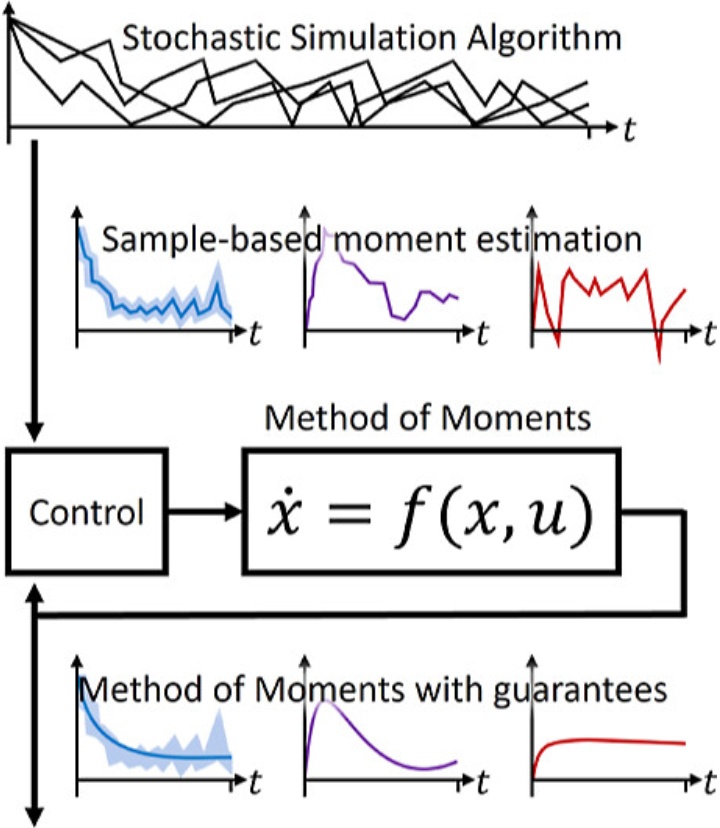
A control system for estimated moments
The Method of Moments is used for describing the average behavior of a system of chemical reactions with a small number of molecules. In practice, this usually involves the expected value and the variance.
The Method of Moments provides a way to reduce the system. Instead of the entire distribution, a manageable number of moments are considered that characterize the distribution well. This means that not the entire distribution is considered, but only the average number of molecules or the variance. In order to set up a consistent system of equations for the moments, the moments must be closed, since lower-order moments generally depend on higher-order moments. Closing means that the dependencies must be interrupted in order to be able to calculate anything at all, as these would otherwise increase to infinity.
In order to be able to apply the Method of Moments, the higher-order moments in the equations must be estimated. To ensure that this estimation doesn’t lead to inaccurate results, Nicole Radde and her team developed the so-called control closure scheme, which is a kind of control system that avoids this problem. Compared to other existing solutions, in this method the calculated moments are guaranteed to remain close to the true moments and cannot deviate arbitrarily. This control closure scheme has great potential for broad application in the description of the moments of biochemical reaction systems.
Read more:
Wagner V, Strässer R, Allgöwer F, Radde N (2023). A provably convergent control closure scheme for the method of moments arising from the Chemical Master Equation, Journal of Chemical Theory and Computation 19(24), 9049–9059, DOI: 10.1021/acs.jctc.3c00548
About the scientist
Nicole Radde holds a Teaching Degree in Mathematics and Physics. In her thesis in the field of theoretical astrophysics and quantum mechanics, she investigated the interactions of neutrinos with nucleons that occur in neutron stars. For her doctoral degree studies, she shifted the focus to the field of life sciences and began to apply her research results to biological systems. Today, she is Professor of Mathematical Modeling and Simulation of Cellular Systems at the University of Stuttgart.