DNA methylation on CG sequences is a chemical modification of DNA that is essential for human embryonic development. The DNA methyltransferase DNMT1 plays a central role in this process. The enzyme uses a spectacular base flipping mechanism, the basics of which are not well understood. An international research team led by Prof. Albert Jeltsch from the Institute for Biochemistry and Technical Biochemistry (IBTB) at the University of Stuttgart used biochemical and structural biological experiments to clarify the dynamic processes involved in this process.
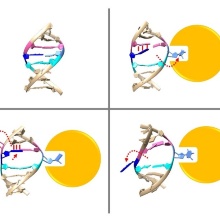
In 1993 it was shown for the first time that DNA methyltransferases partially destroy the DNA double helix structure discovered by Watson and Crick in 1953 in the course of their catalyzed reaction, because the cytosine base to be methylated is flipped out of the DNA helix. The guanine, originally forming a base pair with the cytosine, is left behind in the DNA helix, which leads to dynamic structural changes in the DNA, which, however, have not yet been systematically investigated.
A team led by Prof. Albert Jeltsch from the University of Stuttgart was able to measure the DNA methylation rate of DNMT1 methyltransferase on thousands of DNA sequences containing CG target sequences using a new method. It was shown that the sequence environment has a significant influence on the enzyme activity. In cooperation with a group from the University of California, structures of DNMT1 with different DNA sequences could be solved, which show that the structural changes of the DNA after base flipping depend directly on the neighboring DNA sequence, and these effects also control the turnover rate of DNMT1 . Complexes with minor structural changes showed a high turnover rate, while a complex with a massive structure change showed a slow turnover rate.
With the help of modeling experiments and simulations in cooperation with the group of Prof. Radde from the University of Stuttgart, the mechanism of the reaction could be described in more detail. Further experiments showed that the DNA methylation pattern in human cells and also the effect of DNMT inhibitors used in cancer treatment are strongly influenced by the sequence dependence of DNMT1 activity. "It is fascinating to see how dynamic processes in biomolecules at the atomic level are reflected in global properties such as genome-wide DNA methylation patterns in human cells and the effects of drugs," says Jeltsch.